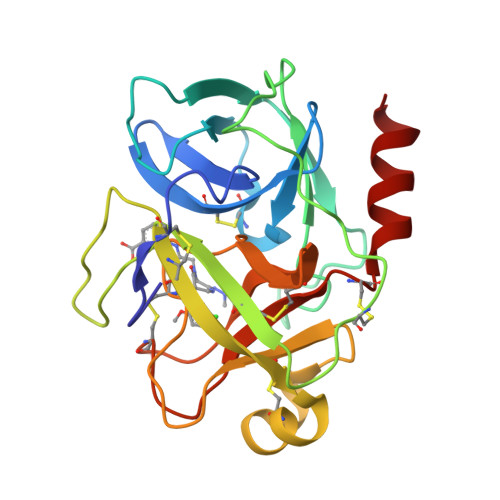
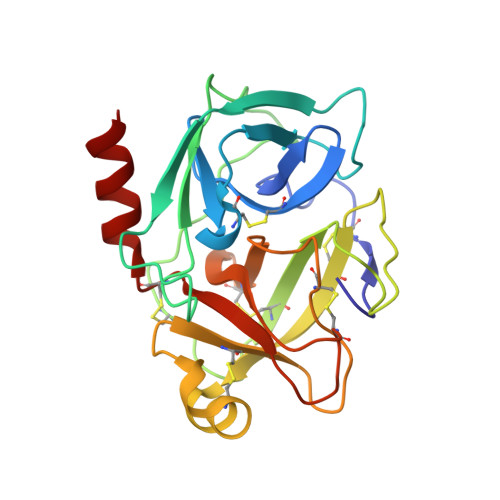
Discovery and structure-activity relationship study of 1,3,6-trisubstituted 1,4-diazepane-7-ones as novel human kallikrein 7 inhibitors
Murafuji, H., Sakai, H., Goto, M., Imajo, S., Sugawara, H., Muto, T.(2017) Bioorg Med Chem Lett 27: 5272-5276
- PubMed: 29102227
- DOI: https://doi.org/10.1016/j.bmcl.2017.10.030
- Primary Citation of Related Structures:
5Y9L, 5YJK - PubMed Abstract:
Compound 1, composed of a 1,3,6-trisubstituted 1,4-diazepane-7-one, was discovered as a novel human kallikrein 7 (KLK7, stratum corneum chymotryptic enzyme, SCCE) inhibitor, and its derivatives were synthesized and evaluated. Structure-activity relationship studies of the amidoxime unit and benzoic acid part of this new scaffold led to the identification of 25 and 34, which were more potent than the hit compound, 1. The X-ray co-crystal structure of compound 25 and human KLK7 revealed the characteristic interactions and enabled explanations of the structure-activity relationship.
Organizational Affiliation:
Asubio Pharma Co., Ltd., 6-4-3 Minatojima-Minamimachi, Chuo-ku, Kobe, Hyogo 650-0047, Japan. Electronic address: murafuji.hidenobu.bc@asubio.co.jp.